Defining Cell Clusters
To reproduce the internal zones defined in the gPROMS network model, you divide the Simcenter STAR-CCM+ mesh into cell clusters.
To enable cell clustering, you require an additional model in the physics continuum for the fluid simulation in Simcenter STAR-CCM+:
- In the relevant physics continuum, activate the Coordinate Cell Clustering model.
When creating clusters, you specify the number of divisions
along each axis of the chosen coordinate system. Two method are provided by which
you specify the number of divisions:
- Number of Subdivisions — allows you to define an equal clustering distribution in all three axes. The subdivision points are spaced equally along each axis.
- Control Points — allows you to define a non-equidistant clustering distribution for each axis. A linear parameter specifies the clustering which fits the control points to the bounding box of the domain being clustered.
- To set the number of divisions, right-click the node and select either Number of Subdivisions or Control Points.
-
Select the node and set the Cluster Merging
Threshold.
For more information see Coordinate Cell Clustering Solver.
-
Select the node and set the following properties:
- Coordinate System: Select the coordinate system with respect to which the mesh is divided into clusters. Depending on the domain symmetries, you can use either a Cartesian, cylindrical, or spherical coordinate system.
- When using the Subdivision option, set Subdivisions to the number of equal portions into which the mesh is divided in each coordinate direction.
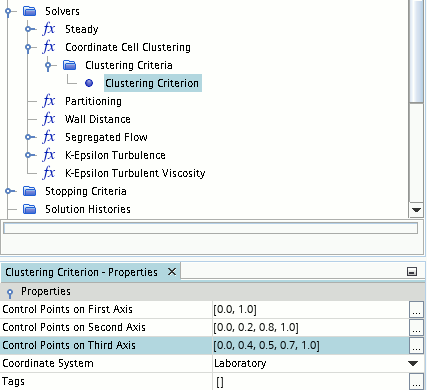
- When using the Control Points option, for each of the three
axes (first, second, and third), provide a vector list of the control
point divisions along the axis. The vector list need not contain actual
measured distances; Simcenter STAR-CCM+
normalizes the divisions according to the bounding dimensions of the
domain in each direction. As a minimum the list must contain the first
and last control point divisions which by default are 0 and 1
(corresponding to no divisions). Any unordered lists are ordered
automatically. For an example see the following screenshot:
For more information, see Coordinate Cell Clustering.
To divide the mesh into clusters:
- Run the Coordinate Cell Clustering Solver for at least one iteration.
-
To visualize the clusters, you can plot the Cell Cluster
Index field function in a scalar scene.
For more information, see Coordinate Cell Clustering.